 |
 |
 |
 |
 |
 T Cell Enumeration and Procedure for T Cell Depletion
T Cell Enumeration and Procedure for T Cell Depletion
Dr. Lawrence S. Lamb, Jr. Richland Memorial Hospital
University of South Carolina School of Medicine
PRINCIPLE
The elimination of T lymphocytes from bone marrow grafts is done to aid in the prevention of graft versus host disease (GVHD). Mononuclear cells from the donor bone marrow are incubated with an antibody (OKT3), followed by incubation with complement. Cells possessing the CD3 antigen (T lymphocytes) are coated with the antibody and undergo complement mediated cytotoxicity. The antibody negative cells (CD3 negative) are washed and prepared for infusion into the bone marrow patient. Flow cytometric detection of CD3, CD5, and CD34+ cells "PRE" and "POST" T cell depletion is utilized in order to calculated the amount of T cells (CD 3+) have been removed from the graft. The use of 7-aminoactinomycin D (7-AAD) is used to discriminate dead cells from live cells by staining the DNA in cells which have lost their integrity of their cellular membrane. The goal of the immunophenotypic analysis of T-cell depleted bone marrow grafts is to estimate the level of depletion. This procedure describes the methods used to stain three samples of the donor bone marrow graft: before, during, and after T-cell depletion. In addition to determining the viability of the pre- and post-T cell depletion procedures.
PRECAUTION: SPECIMENS SHOULD BE HANDLED ACCORDING TO LOCAL EXPOSURE CONTROL PLANS AND OSHA’S BLOOD BORNE PATHOGEN STANDARDS.
SPECIMEN: Fresh harvest, Pre, and Post T-cell depleted marrow obtained from Marrow
Processing Laboratory.
REAGENTS AND EQUIPMENT:
1. Selected antibodies (See Table 1).
2. 12 X 75 polypropylene test tubes.
3. 12 X 75 test tube racks.
4. Rainin multipipetter.
5. 10mL pipette and Pipette aid.
6. Plug caps.
7. Parafilm.
8. 15 mL conical polypropylene test tubes.
9. Centrifuge
10. Domestic refrigerator (1-6° C).
11. Phosphate buffered saline (Sigma D-8537).
12. 1% Formaldehyde Fixative (Mallinckrodt 5016).
13. Vortex mixer (Maxi-mix II).
14. FACS Lyse solution (Becton-Dickinson 92-0002)
PROCEDURE:
For "FRESH", "PRE" and "POST"-T cell Depletions:
1.1. Prepare 12 X 75 polypropylene tubes for T-cell depletion panel (See Table 1).
1.2. Add 10m l of the appropriate antibody.
1.3. Obtain samples from Marrow Processing Laboratory.
1.4. Record nucleated counts on appropriate panel sheets.
1.5. Record and cross reference sample accession number.
1.6. Add 100m l of marrow sample to each tube.
1.7. Centrifuge for 15 seconds at 400 g and vortex (This ensures mixing of cells and reagents).
1.8. Incubate "FRESH" sample at ambient temperature for 30 minutes in the dark. For "PRE" and "POST"-T cell depletion samples, incubate for 20 minutes at 4° C in domestic refrigerator.
1.9. Only to the "FRESH" sample, add 2 ml of FACS Lyse solution and incubate at ambient temperature for 7-10 minutes in the dark..
1.10. Add 2 mL PBS and spin 5 minutes at 600 g.
1.11. Remove supernatant and add 200 m l fixative to all tubes.
1.12. Acquire on flow cytometer (The "PRE" and "POST" samples are live gated on the CD45+ cell population in order to exclude debris.) Immediately proceed with analysis of sample.
1.13. Return results to Marrow Processing Laboratory STAT. After obtaining the director’s, or designee’s, signature on the report.
For "POST"-T cell Depletion Samples:
2.1. Add 10 ml of PBS to the post-T cell depletion sample when received from the Marrow Processing Laboratory.
2.2. Spin at 800 g for 5 minutes.
2.3. Decant PBS and resuspend cell pellet in 1 ml of PBS.
2.4. Incubate sample at 37oC for 60 minutes.
2.5. After incubation, obtain a nucleated cell count using a hematology analyzer.
2.6. Proceed with the above procedure for staining..
PROCEDURE NOTES:
The T-cell depletion panel described in Table 1 is stained using the following samples: The "FRESH HARVEST" sample is merely a diluted sample straight from the bone marrow bag. The "PRE" sample is obtained from the Stericell. The "POST" sample is after complement incubation.
LIMITATIONS
:· Due to the nature of these samples (reduced cellularity and limited volume), a restricted panel of monoclonal antibodies is used for analysis.
· Monoclonal antibodies reacting with granulocytes may require additional amounts of
antibodies to assure saturation of all binding sites.
· An extension of the unstained cluster in the control tube(s) may be seen if Fc binding occurs (usually due to an excess of monoclonal antibody). Fc receptor binding monoclonal antibodies is of lower avidity than is specific marker binding.
· Monoclonal reagents against a cell surface antigen or receptor which is occupied by plasma components can have reduced staining intensity when analyzed with lysed whole blood methodology.
· Samples with clumped platelets, may have falsely high CD19 values, due to platelets staining with CD19.
QUALITY CONTROL:
As daily quality control of several monoclonal antibodies, commercial controls are run on the flow cytometer. As each new lot of a particular antibody is used, a parallel tube is run to assure consistent reactivity.
ANALYSIS
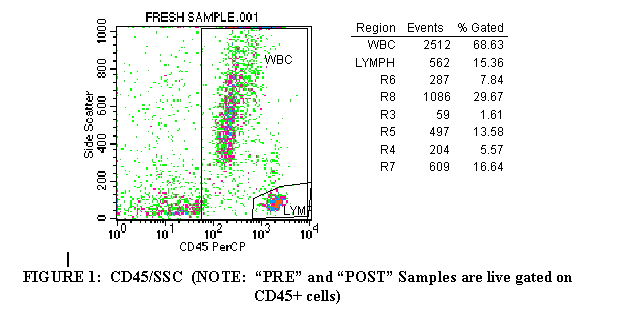
3.1 Import the CD45 PRCP/SSC plot of the first tube (Figure 1).
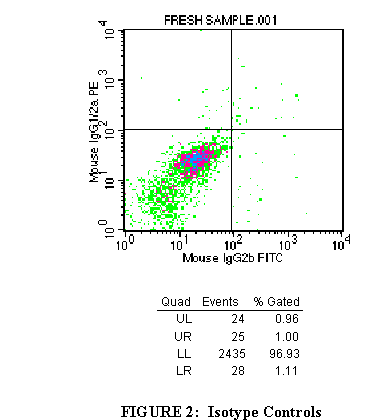
3.2 Import the FL1/FL2 plot of the first tube into the top right window
(Figure 2).
3.4 Click on the polygon symbol from the palette to the left of the document.
Draw a region around the CD45+ cells in order to exclude debris and RBC’s from the analysis gate. Label this region "WBC" (Figure 1).
The CD45-Hle-1, sometimes known as the Leukocyte common antigen,
is expressed by all nucleated WBC and most WBC progenitors in blood and bone marrow.
3.5 Click on the polygon symbol from the palette to the left of the document. Draw a region around the lymphocyte population (Figure 1). Label this
region "LYMPH". Lymphocytes have scatter properties less granular than monocytes or granulocytes, thus their placement in the lower right corner of the plot.
3.6 Import the CD45 PRCP/SSC plot of the second tube to the second
left window. Continue importing plots as described in steps 3.2 through
3.3 until all plots are filled. Draw quadrant markers for the plots (See 1.8)
3.7 Under the "STATISTICS" menu, select "region stats" and "quadrant stats" for each plot.
3.8 Calculate the percentage of lymphocytes in the CD45+ population by dividing "LYMPH" by "WBC" and multiplying by 100.
3.9 DETERMINATION OF CD3+, CD5+, CD34+ CELLS
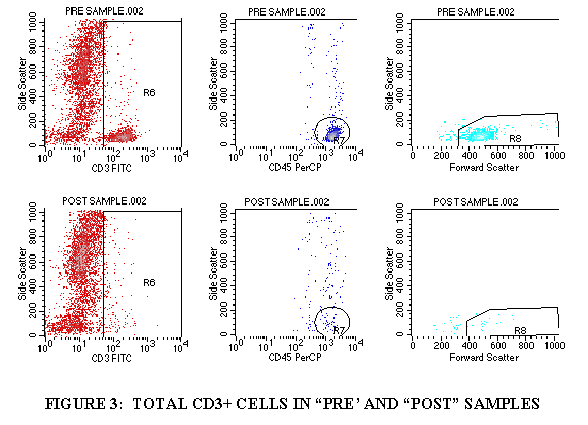
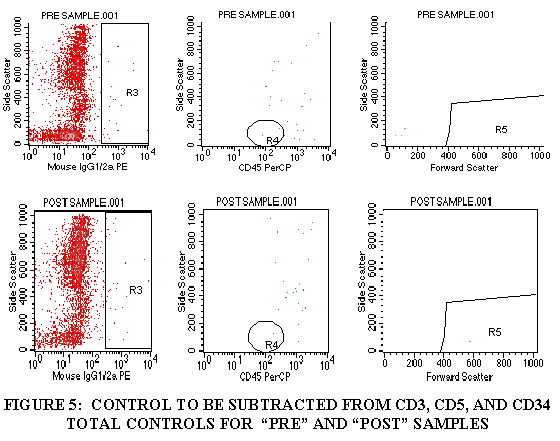
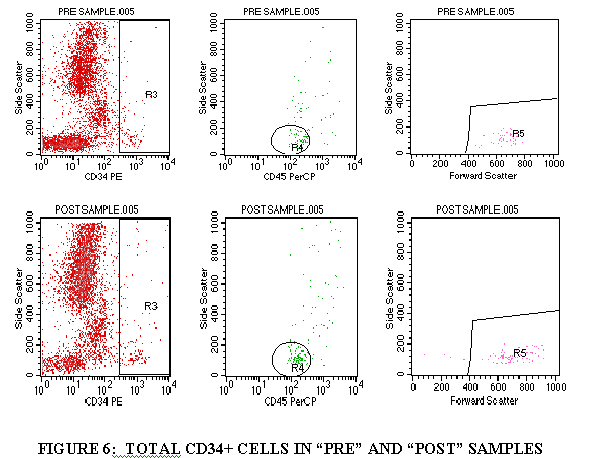
3.9.1 Import FL1/SSC of the tube containing CD3 (through the WBC gate) (Figures 3).
3.9.2 Draw a region aroung the CD3+ cell population.
3.9.3 Import the FL3/SSC of the tube containing CD3 (through the CD3+ gate and the WBC gate) (Figure 3).
3.9.4 Draw a region around the CD45+ cells in this plot (Figure 3).
3.9.5 Import the FSC/SSC of the tube containing CD3 (through the CD3+ gate, the WBC gate, and the CD45+ gate from Step 3.9.4) (Figure 3).
3.9.6 Draw a region around the FSC+ cell population (Figure 3).
3.9.7 Divide the FSC+ cells by the WBC population to get the TOTAL CD3+ cells.
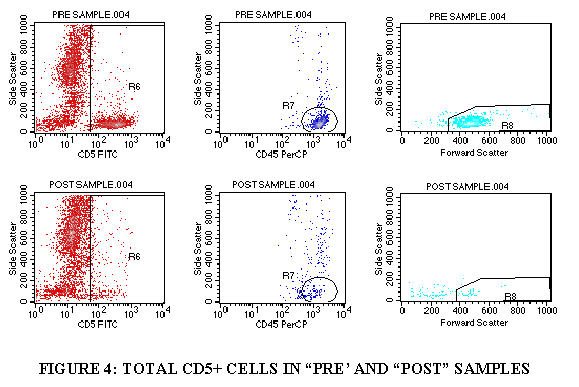
3.9.8 Repeat steps 3.9.1 through 3.9.7 for the control tube, CD5 tube and the CD34 tube (Figures 4-6).
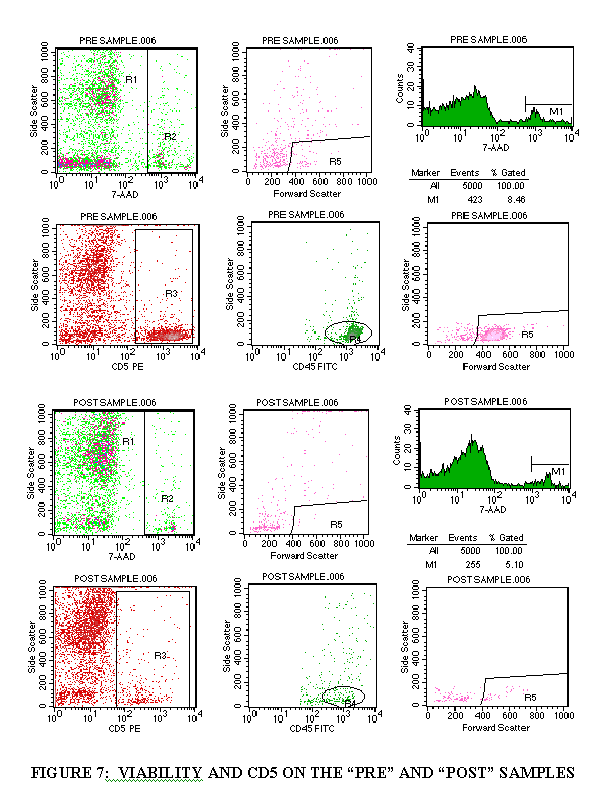
3.10 DETERMINATION OF CELL VIABILITY AND CD5+ CELLS
3.10.1 Import the FL3/SSC plot of the viability tube (Figure 7).
3.10.2 Draw a region around the "dead" cell population (Figure 7).
3.10.3 Import the FSC/SSC plot of the viability tube (through the "dead" cell population) (Figure 7).
3.10.4 Draw a region around the FSC+ cells (Figure 7).
3.10.5 Import a 7-AAD/SSC histogram plot (no gate) (Figure 7).
3.10.6 Draw a marker over the 7-AAD+ population (Figure 7).
3.10.7 Subtract the 7-AAD+ population from 100 to get the viability percentage.
3.10.8 Import the gating as described above (steps 3.9.1 through 3.9.7) to get the CD5 total and the viable CD5+ cells.
REFERENCES:
Becton Dickinson Immunocytometry Systems. (1993): Simultest package insert. Publishing
# 4520, 110.
Fritsch, G., Emminger, W., Buchinger, P., Printz, D., and Gadner, H. (1991): CD-34 Positive Cell
Proportions in Peripheral Blood Correlate with Colony Forming Capacity. Exp. Hematol.
19:1079.
Gee, A. (1993): Preparation of samples for Flow Cytometric Analysis, p. 1-4. In Leon W.
M.M. Terstappen (course director), ISHAGE Training Courses-Multidimensional Flow
Cytometric Analysis of Bone Marrow & Peripheral Blood Stem Cells. Becton Dickinson
Immunocytometry Systems, San Jose.
Jackson, A.L., and N.L. Warner. (1986): Preparation,Staining, and Analysis by Flow Cytometry of Peripheral Blood Leukocytes, p.226-227. In N.R. Rose (ed), Manual of Clinical
Laboratory Immunology. American Society for Microbiology, Washington, D.C.
Kuby, Janis (1992): Cells and Organs of the Immune System, p. 54-58. In Immunology.
W.H. Freeman and Company. New York.
Melamed, M., Mullaney, P., Menelsohn, M., eds. (1979):Flow Cytometry and Sorting. John Wiley, New York.
Parks,D.R., L.L.Lanier, and L.A. Herzenberg. (1985): Flow Cytometry and fluorescence activated cell sorting (FACS), p.107-117. In D.M. Weir (ed), Handbook of Experimental Immunology. C.V. Mosby Co., St.Louis.
Reinherz, E.L., B.F. Haynes, L.M. Nadler, and I.D. Bernstein (ed). 1985. Leucocyte Typing, vol. 2. Springer-Verlag, New York.
Shapiro, Howard M., (1995): Parameters and Probes, Practical Flow Cytometry, 3rd Edition , Wiley-Liss, New York.
Steinkamp, J.A. 1984. Flow Cytometry. Rev. Sci. Instrum. SS:1375-1400.Stewart, CC, and Stewart, SJ, (1997): Immunophenotyping, Current Protocols in Flow Cytometry, John Wiley and Sons, New York
Wunder, E., Sovalat, H., Fritsch, G., Silvestri, F., Henon, P, and Serke, S. (1992): Report on the European Workshop on peripheral blood stem cell determination and standardization. J.
Hematotherapy, 1:131.







 |
 |
 |
 |
 |
CD-ROM Vol 3 was produced by Monica M. Shively and other staff at the Purdue University Cytometry Laboratories and distributed free of charge as an educational service to the cytometry community. If you have any comments please direct them to Dr. J. Paul Robinson, Professor & Director, PUCL, Purdue University, West Lafayette, IN 47907. Phone:(765) 494-0757; FAX (765) 494-0517; Web http://www.cyto.purdue.edu, EMAIL cdrom3@flowcyt.cyto.purdue.edu